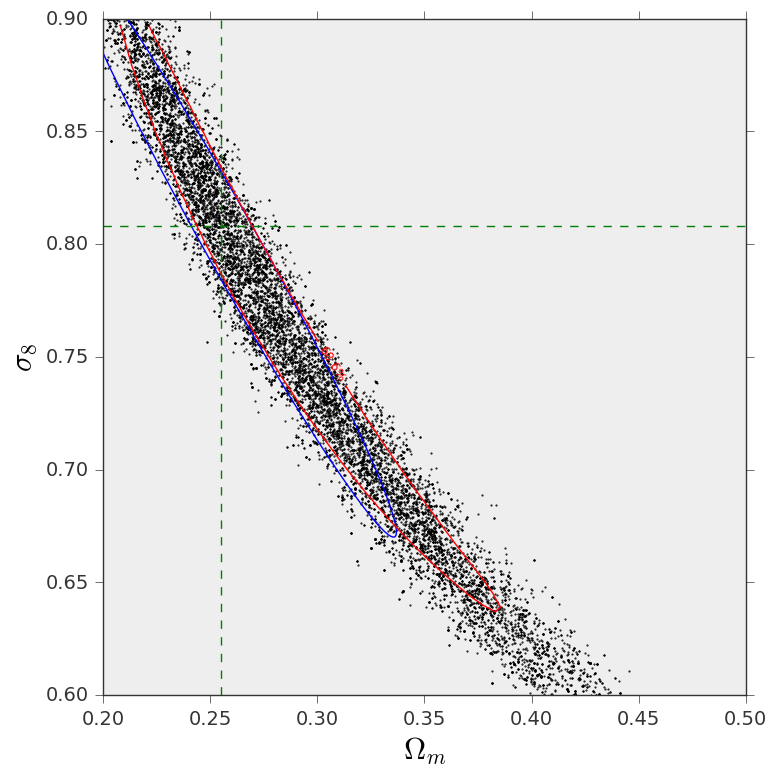
Three different ways to do parameter sampling¶
This code snipped shows how to use LensTools to perform cosmological parameter sampling with three diffenent methods: direct evaluation of the likelihood, Fisher Matrix and MCMC
from lenstools.statistics.ensemble import Series,Ensemble
from lenstools.statistics.constraints import Emulator
from lenstools.statistics.contours import ContourPlot
import numpy as np
import matplotlib.pyplot as plt
def lt_sample(emulator,test_data,covariance,p_value=0.68):
#Check that the data types are correct
assert isinstance(emulator,Emulator)
assert isinstance(test_data,Series)
assert isinstance(covariance,Ensemble)
#Plot setup
fig,ax = plt.subplots(figsize=(8,8))
#Map the likelihood in the OmegaM-sigma8 plane, fix w to -1
p = Ensemble.meshgrid({
"Om":np.linspace(0.2,0.5,50),
"sigma8":np.linspace(0.6,0.9,50)
})
p["w"] = -1.
#Compute the chi squared scores of the test data on a variety of parameter points
scores = emulator.score(p,test_data,covariance,correct=1000,method="chi2")
scores["likelihood"] = np.exp(-0.5*scores[emulator.feature_names[0]])
contour = ContourPlot.from_scores(scores,parameters=["Om","sigma8"],
feature_names=["likelihood"],
plot_labels=[r"$\Omega_m$",
r"$\sigma_8$"],fig=fig,ax=ax)
contour.getLikelihoodValues([p_value],precision=0.01)
contour.plotContours(colors=["red"])
contour.labels()
#Approximate the emulator linearly around the maximum (Fisher matrix)
fisher = emulator.approximate_linear(center=(0.26,-1.,0.8))
#Consider (OmegaM,sigma8) only
fisher.pop(("parameters","w"))
fisher = fisher.iloc[[0,1,3]]
#Fisher confidence ellipse
ellipse = fisher.confidence_ellipse(covariance,
correct=1000,
observed_feature=test_data,
parameters=["Om","sigma8"],
p_value=p_value,
fill=False,
edgecolor="blue")
ax.add_artist(ellipse)
#MCMC sampling of (OmegaM,sigma8)
samples = emulator.sample_posterior(test_data,
features_covariance=covariance,
correct=1000,
pslice={"w":-1},
sample="emcee")[emulator.feature_names[0]]
ax.scatter(samples["Om"],samples["sigma8"],marker=".",color="black",s=1)
ax.set_xlim(0.2,0.5)
ax.set_ylim(0.6,0.9)
#Save the figure
fig.tight_layout()
fig.savefig("parameter_sampling.png")
And this is the resulting figure: